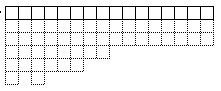
 The GAArray object
The GAArray objectThe squares are elements in the array. Arrays are 1 dimensional, but derived classes can have 2 or more dimensions. Each element contains a user-specified object.
This document describes the programming interface for the GA library. The section for each class contains a description of the object's purpose followed by the creator signature and member functions. There are also sections for library constants, typedefs, and function signatures.
see also: library overview, class hierarchy
Table of contents
General library information |
Genomes and related objects |
typedef float GAProbability, GAProb
typedef enum GABoolean {gaFalse, gaTrue} GABool
typedef float (*GAObjectiveFunction)(GAGenome &);
typedef float (*GAVectorObjectiveFunction)(const GAObjectiveVector &);
typedef GABoolean (*GACompletionFunction)(GA & ga);
typedef GAGenome & (*GAReplacementFunction)(GAGenome &, GAPopulation &);
typedef GAGenome & (*GASelectionFunction)(const GAPopulation &);
typedef float (*GADistanceFunction)(GAGenome &, GAGenome &);
typedef void (*GAInitializationOperator)(GAGenome &);
typedef int (*GAMutationOperator)(GAGenome &, float);
typedef void (*GACrossoverOperator)(GAGenome &,
const GAGenome &, const GAGenome &);
typedef void (*GAPopulationInitializer)(GAPopulation &);
typedef void (*GAPopulationEvaluator)(GAPopulation &);
typedef void (*GAPopulationSorter)(GAPopulation &);
const int gaDefNumGen = 250; // number of generations const float gaDefPConv = 0.05; // convergence percentage const int gaDefNConv = 20; // number of generations for convergence const float gaDefPMut = 0.01; // mutation probability const float gaDefPCross = 0.9; // crossover probability const int gaDefNumBestChrom = 1; // number of best genomes to keep track of const int gaDefScoreFrequency1 = 1; // non-overlapping population score frequency const int gaDefScoreFrequency2 = 100; // overlapping population score frequency const int gaDefNumOffspring = 2; // number of offspring const int gaDefPopSize = 30; // population size const float gaDefLinearScalingMultiplier = 1.2; const float gaDefSigmaTruncationMultiplier = 2.0; const float gaDefPowerScalingFactor = 1.0005;
The GARandomSeed function uses the current time multiplied by the process ID (on systems that have PIDs) as an argument to the system's srandom function. On systems with no process IDs it uses only the time. You can specify your own random seed if you like.
The GARandomBit function is the most efficient way to do unbiased coin tosses. It caches bits so that it calls your system's random number generator a minimum number of times.
void GARandomSeed()
void GARandomSeed(const unsigned s)
int GARandomInt()
int GARandomInt(const int low, const int high)
double GARandomDouble()
double GARandomDouble(const double low, const double high)
float GARandomFloat()
float GARandomFloat(const float low, const float high)
int GARandomBit()
GABoolean GAFlipCoin(float p)
The completion function returns gaTrue when the genetic algorithm should finish, and gaFalse when the genetic algorithm should continue.
GABoolean GATerminateUponGeneration(GA &) GABoolean GATerminateUponConvergence(GA &) GABoolean GATerminateUponPopConvergence(GA &)
Convergence is a number between 0 and 1. A convergence of 0 means that the nth previous best-of-generation is equal to the current best-of-generation. When you use convergence as a stopping criterion you must specify the convergence percentage and you may specify the number of previous generations against which to compare. The GA will always run at least this many generations.
GAGenome & GARankSelector(const GAPopulation &) GAGenome & GARouletteWheelSelector(const GAPopulation &) GAGenome & GATournamentSelector(const GAPopulation &) GAGenome & GADSSelector(const GAPopulation &) GAGenome & GASRSSelector(const GAPopulation &) GAGenome & GASUSSelector(const GAPopulation &)
If you specify Custom or Crowding replacement then you must also specify a replacement function. The replacement function takes as arguments an individual and the population into which the individual should be placed. It returns a reference to the genome that the individual should replace. If the individual should not be inserted into the population, the function should return a reference to the individual.
Not every replacement method can be used with every genetic algorithm object. See the documentation for the specific type of genetic algorithm for more details.
GAEvery GA derived from this class has the following member functions:
void * userData()
void * userData(void *)
GASelectionFunction selectionFunction()
GASelectionFunction selectionFunction(GASelectionFunction)
GACompletionFunction completionFunction()
GACompletionFunction completionFunction(GACompletionFunction)
const GAScalingScheme & scalingScheme()
const GAScalingScheme & scalingScheme(const GAScalingScheme &)
const GAParameters & parameters()
const GAParameters & parameters(const GAParameters &)
int nGenerations()
int nGenerations(const unsigned int)
float pMutation()
float pMutation(const float)
float pCrossover()
float pCrossover(const float)
int populationSize()
int populationSize(const unsigned int)
float pConvergence()
float pConvergence(const float)
int nConvergence()
int nConvergence(const unsigned int)
int nBestGenomes(const unsigned int n = gaDefNumBestChrom)
int scoreFrequency(const unsigned int f = 1)
const GAStatistics & statistics()
GAPopulation & population()
GAGenome & bestOfPop(unsigned int n = 0)
GAGenome & bestOfAll(unsigned int n = 0)
void scores(ostream & outputstream)
void scores(const char * filename)
float convergence()
int generation()
void initialize(const unsigned int seed)
void evolve()
void step()
int done()
If you evolve the GA without using the evolve member function, be sure to call initialize before stepping through the evolution. You can use the step member function to evolve a single generation. The evolve member function simply calls the step member function until the done member function returns gaTrue.
- userData
- Set/Get the userData member of the genetic algorithm. This member is a generic pointer to any information that needs to be stored with the genetic algorithm.
- selectionFunction
- Set/Get the selection function for the genetic algorithm. The selection function is used to pick individuals from a population before mating and mutation occur. The function must have the proper signature.
- completionFunction
- Set/Get the termination function. The genetic algorithm is complete when the completion function returns gaTrue. The function must have the proper signature.
- scalingScheme
- Set/Get the scaling scheme. The specified scaling scheme must be derived from the GAScalingScheme class.
- parameters
- Set/Get the parameters for the genetic algorithm. Use this member function to set the population size, number of generations, mutation and crossover probabilities, score frequency, and number of best genomes to record. A score frequency of 10 means that the best-of-generation will be recorded every 10th generation. A score frequency of 0 means that the scores will not be recorded.
- nGenerations
- Set/Get the number of generations.
- pMutation
- Set/Get the mutation probability.
- pCrossover
- Set/Get the crossover probability.
- populationSize
- Set/Get the population size.
- pConvergence
- Set/Get the convergence percentage. The convergence is defined as the ratio of the Nth previous best-of-generation score to the current best-of-generation score. N is defined by the nConvergence member function.
- nConvergence
- Set/Get the number of generations used for the convergence test.
- nBestGenomes
- Specify the number of unique best genomes to keep track of. The default is 1. Notice that the more you keep track of, the slower the GA will run. This is because at each generation the GA must compare all the 'best' genomes to the best in the current population.
- scoreFrequency
- Specify how often the best-of-generation score should be recorded. The default depends on the type of genetic algorithm that you are using.
- statistics
- Returns a reference to the statistics object in the genetic algorithm.
- population
- Returns a reference to the current population.
- bestOfPop
- Returns a reference to the best individual in the current population.
- bestOfAll
- Returns a reference to the best individual encountered since the genetic algorithm was initialized.
- scores
- Dump the best-of-generation objective (not fitness) scores to stdout or to a file (depending on how you call it).
- convergence
- Returns the current convergence. The convergence is defined as the ratio of the Nth previous best-of-generation score to the current best-of-generation score.
- generation
- Returns the current generation.
- initialize
- Initialize the genetic algorithm. If you specify a seed, this function calls GARandomSeed with that value. It then initializes the population and does the first population evaluation.
- evolve
- Initialize the genetic algorithm then evolve it until the termination criteria have been satisfied.
- step
- Evolve the genetic algorithm for one generation.
- done
- Returns gaTrue if the termination criteria have been met, returns gaFalse otherwise. This function simply calls the completion function that was specified using the completionFunction member function.
Elitism is optional. By default, elitism is on. To turn off elitism, pass gaFalse to the elitist member function. The score frequency for this GA defaults to 1 (it records the best-of-generation every generation). The default scaling is Linear, the default selection is RouletteWheel. The increment operator (++) increments the GA by one generation.
GASimpleGA(const GAGenome &, void * userData = NULL)
GASimpleGA(const GAPopulation &, void * userData = NULL)
void objectiveVector(const GAObjectiveVector & v)
void objectiveFunction(GAObjectiveFunction)
GASimpleGA & operator++()
GABoolean elitist()
GABoolean elitist(const GABoolean flag)
see also: GA base class
You can select the amount of overlap between generations by specifying the pReplacement parameter. This is the percentage of the population that should be replaced each generation. The worst members are replaced with the newly generated offspring (even if the new ones are less fit than the originals).
Elitism is optional. If you specify a replacement rate less than 100% then the elitism flag is ignored. The default is gaTrue. The score frequency for this GA defaults to 100 (it records the best-of-generation every 100th generation). The default scaling is Linear, the default selection is RouletteWheel. The increment operator (++) increments the GA by one generation.
Use the replacementScheme method to specify which type of replacement the GA should use. This GA works with all of the replacement schemes except for gaReplaceParent. If you specify gaReplaceCustom or gaReplaceCrowding you must also specify a replacement function with the proper signature.
GASteadyStateGA(const GAGenome &, void * userData = NULL)
GASteadyStateGA(const GAPopulation &, void * userData = NULL)
void objectiveVector(const GAObjectiveVector & v)
void objectiveFunction(GAObjectiveFunction)
GASteadyStateGA & operator++()
float pReplacement() const
float pReplacement(const float percentage)
GABoolean elitist()
GABoolean elitist(const GABoolean flag)
GAReplacementScheme replacementScheme()
GAReplacementScheme replacementScheme(GAReplacementScheme,
GAReplacementFunction f = NULL
see also: GA base class
You can specify the number of children that are generated in each 'generation' by using the nOffspring member function. Since this GA is based on a two-parent crossover model, the number of offspring must be either 1 or 2. The default is 2. The score frequency for this GA defaults to 100 (it records the best-of-generation every 100th generation). The default scaling is Linear, the default selection is RouletteWheel. The increment operator (++) increments the GA by one generation.
Use the replacementScheme method to specify which type of replacement the GA should use. This GA works with all of the replacement schemes. If you specify gaReplaceCustom or gaReplaceCrowding you must also specify a replacement function with the proper signature.
GAReplacementGA(const GAGenome &, void * userData = NULL)
GAReplacementGA(const GAPopulation &, void * userData = NULL)
void objectiveVector(const GAObjectiveVector & v)
void objectiveFunction(GAObjectiveFunction)
GAReplacementGA & operator++()
GAReplacementScheme replacementScheme()
GAReplacementScheme replacementScheme(GAReplacementScheme,
GAReplacementFunction f = NULL
int nOffspring() const
int nOffspring(const unsigned int n)
see also: GA base class
GAObjectiveVector(unsigned int n,
GAVectorObjectiveFunction function=GAObjectiveProduct)
int size() const
int resize(const unsigned int i)
GAVectorObjectiveFunction evaluator() const
GAVectorObjectiveFunction evaluator(GAVectorObjectiveFunction f)
float component(const unsigned int i) const
float component(const unsigned int i, const float f)
const float & operator[](const unsigned int i) const
float & operator[](const unsigned int i)
float evaluate() const
GAObjectiveVector & operator+=(const float value)
GAObjectiveVector & operator-=(const float value)
GAObjectiveVector & operator*=(const float value)
GAObjectiveVector & operator/=(const float value)
GAObjectiveVector & operator+=(const GAObjectiveVector & orig)
GAObjectiveVector & operator-=(const GAObjectiveVector & orig)
GAObjectiveVector & operator*=(const GAObjectiveVector & orig)
GAObjectiveVector & operator/=(const GAObjectiveVector & orig)
friend int operator==(const GAObjectiveVector &a, const GAObjectiveVector &b)
friend int operator!=(const GAObjectiveVector &a, const GAObjectiveVector &b)
friend int operator<(const GAObjectiveVector &a, const GAObjectiveVector &b)
friend int operator>(const GAObjectiveVector &a, const GAObjectiveVector &b)
friend int operator<=(const GAObjectiveVector &a, const GAObjectiveVector &b)
friend int operator>=(const GAObjectiveVector &a, const GAObjectiveVector &b)
Here are the built-in objective evaluation functions. Use the evaluator member function to change the evalution function. You can define your own evaluator, but it must have the proper signature.
float GAObjectiveProduct(const GAObjectiveVector &) float GAObjectiveSummation(const GAObjectiveVector &)
enum GADimension { gaLength, gaWidth, gaHeight, gaDepth }
enum GACloneMethod { gaCloneContents, gaCloneAttributes }
The dimension is used to specify which dimension is being referred to in multi-dimensional genomes. The clone method specifies whether to clone the entire genome (a new genome with contents identical to the original will be created) or just the attributes of the genome (a new genome with identical characteristics will be created). In both cases the caller is responsible for deleting the memory allocated by the clone member function.
Every genome class inherits the following methods from the base class:
void copy(const GAGenome & c)
GAGenome * clone(const GACloneMethod flag = gaCloneContents)
float score()
float score(const float s)
int mutations()
GA * geneticAlgorithm() const
GA * geneticAlgorithm(GA &)
void * userData() const
void * userData(void * data)
GAObjectiveFunction objectiveFunction() const
GAObjectiveFunction objectiveFunction(GAObjectiveFunction f)
GAObjectiveVector * objectiveVector() const
GAObjectiveVector * objectiveVector(const GAObjectiveVector &)
void initialize()
GAInitializationOperator initializationOperator() const
GAInitializationOperator initializationOperator(GAInitializationOperator)
int mutate(const float pmutation)
GAMutationOperator mutationOperator() const
GAMutationOperator mutationOperator(GAMutationOperator)
void cross(const GAGenome &, const GAGenome &)
GACrossoverOperator crossoverOperator() const
GACrossoverOperator crossoverOperator(GACrossoverOperator op)
GACrossSite & crossoverSite() const
GACrossSite & crossoverSite(const GACrossSite &)
void crossover(GACrossoverOperator, const GACrossSite &)
void mirrorCrossSite(const GAGenome &)
void pickCrossSite()
void pickCrossSite(const GACrossSite &)
void pickCrossSite(const GAGenome &, const GAGenome &)
The clone method allocates space for a new genome whereas the copy method uses the space of the genome to which it belongs. The score member function returns the objective score of the genome using the objective function assigned to the genome. If no objective function has been assigned, a score of 0 will be returned. If the score function is called with an argument, the genome's objective score is set to that value.
The various pick members are used by the genetic algorithm to select a crossover site in the genome. If you call pick with no arguments, a site is selected at random. If you call pick with a crossover site as the argument, that site is copied. If you call pick with two genomes as arguments, a site is selected at random using information in those genomes (this method assumes that the genomes will be the parents for a subsequent crossover operation).
The mirror function picks a 'mirror image' of the crossover site based on the site that has been selected in the genome in the argument to the mirror function. This function is used when two offspring are produced in a crossover of two parents.
You must define an initialization operator for this class. The default initializer is GANoInitializer - if you do not assign an initialization operator then you'll get errors about no initializer defined when you try to initialize the genome.
By default, no mutator or crossover is defined. If you want to force a default mutator, crossover, and initializer, copy the list creator template from the creators.C file and modify it to get the operators you want. Alternatively you can use the initializationOperator, mutationOperator, and crossoverOperator member functions to assign the operators. See the comments in creators.C and the examples for more details.
The library has some built-in mutation and crossover operators. They are defined in list.op.C. If you use these operators you must include list.op.h and define the LIST_TYPE macro to the type that you are going to instantiate. See the list example source code for more details about the limitations of the LIST_TYPE macro and built-in list operators.
GAListGenome(GAObjectiveFunction objective = NULL,
void * userData = NULL,
GA * ga = NULL)
see also: GAListYou must define an initialization operator for this class. The default initializer is GANoInitializer - if you do not assign an initialization operator then you'll get errors about no initializer defined when you try to initialize the genome.
By default, no mutator or crossover is defined. If you want to force a default mutator, crossover, and initializer, copy the list creator template from the creators.C file and modify it to get the operators you want. Alternatively you may use the initializationOperator, mutationOperator, and crossoverOperator member functions to assign the operators. See the comments in creators.C and the examples for more details.
The library has some built-in mutation and crossover operators. They are defined in tree.op.C. If you use these operators you must include tree.op.h and define the TREE_TYPE macro to the type that you are going to instantiate. See the tree example source code for more details about the limitations of the TREE_TYPE macro and built-in tree operators.
GATreeGenome(GAObjectiveFunction objective = NULL,
void * userData = NULL,
GA * ga = NULL)
see also: GATreeThe allele set defines the possible values that each element in the string may assume.
GAStringGenome(const unsigned int length,
GAStringAlleleSet &,
GAObjectiveFunction objective = NULL,
void * userData = NULL,
GA * ga = NULL)
see also: GA1DArrayGenomeThe phenotype defines how bits should map into decimal values and vice versa. A single binary-to-decimal phenotype contains the number of bits used to represent the decimal value and the minimum and maximum decimal values to which the set of bits will map.
GABin2DecGenome(const GABin2DecPhenotype &,
GAObjectiveFunction objective = NULL,
void * userData = NULL,
GA * ga = NULL)
GABin2DecPhenotype & phenotypes(const GABin2DecPhenotype &)
GABin2DecPhenotype & phenotypes() const
int nPhenotypes() const
float phenotype(const unsigned int n) const
float phenotype(const unsigned int n, const float value)
see also: GA1DBinaryStringGenome
GA1DBinaryStringGenome(const unsigned int x,
GAObjectiveFunction objective = NULL,
void * userData = NULL,
GA * ga = NULL)
short gene(const unsigned int x = 0) const
short gene(const unsigned int, const short)
int length() const
int length(const int l)
int resize(const int x)
int resizeBehaviour() const
int resizeBehaviour(const unsigned int minx, const int maxx)
void copy(const GA1DBinaryStringGenome &,
const unsigned int xdest, const unsigned int xsrc, unsigned int length)
see also: GAGenome
GA2DBinaryStringGenome(const unsigned int x, const unsigned int y,
GAObjectiveFunction objective = NULL,
void * userData = NULL,
GA * ga = NULL)
short gene(const unsigned int x, const unsigned int y) const
short gene(const unsigned int x, const unsigned int y, const short value)
int width() const
int width(const int w)
int height() const
int height(const int h)
int resize(const int x, const int y)
int resizeBehaviour(const GADimension which) const
int resizeBehaviour(const GADimension which, const unsigned int min, const int max)
int resizeBehaviour(const unsigned int minx, const int maxx,
const unsigned int miny, const int maxy)
void copy(const GA2DBinaryStringGenome &,
const unsigned int xdest, const unsigned int ydest,
const unsigned int xsrc, const unsigned int ysrc,
const unsigned int width, const unsigned int height);
see also: GAGenome
GA3DBinaryStringGenome(const unsigned int x, const unsigned int y, const unsigned int z,
GAObjectiveFunction objective = NULL,
void * userData = NULL,
GA * ga = NULL)
short gene(const unsigned int x, const unsigned int y, const unsigned int z) const
short gene(const unsigned int x, const unsigned int y, const unsigned int z, const short value)
int width() const
int width(const int w)
int height() const
int height(const int h)
int depth() const
int depth(const int d)
int resize(const int x, const int y, const int z)
int resizeBehaviour(const GADimension which) const
int resizeBehaviour(const GADimension which, const unsigned int min, const int max)
int resizeBehaviour(const unsigned int minx, const int maxx,
const unsigned int miny, const int maxy,
const unsigned int minz, const int maxz)
void copy(const GA3DBinaryStringGenome &,
const unsigned int xdest, const unsigned int ydest, const unsigned int zdest,
const unsigned int xsrc, const unsigned int ysrc, const unsigned int zsrc,
const unsigned int width, const unsigned int height, const unsigned int depth);
see also: GAGenomeBy default, no mutator or crossover is defined. If you want to force a default mutator, crossover, and initializer, copy the list creator template from the creators.C file and modify it to use the operators you want. If you do not use this method then you must use the initializationOperator, mutationOperator, and crossoverOperator member functions to assign the operators. See the comments in creators.C for more details.
When you define an allele set for an array genome, the genome does not make its own copy. Therefore, if you later change the allele set, the array genome will see the changes as well. It will not automatically re-initialize itself.
Use the resizeBehaviour and resize member functions to control the size of the genome. The default behaviour is fixed size. Using the resizeBehaviour method you can specify minimum and maximum values for the size of the genome. If you specify minimum and maximum as the same values then fixed size is assumed. If you use the resize method to specify a size that is outside the bounds set earlier using resizeBehaviour, the bounds will be 'stretched' to accomodate the value you specify with resize. Conversely, if the values you specify with resizeBehaviour conflict with the genome's current size, the genome will be resized to accomodate the new values.
When resizeBehaviour is called with no arguments, it returns the maximum size if the genome is resizable, or gaNoResize if the size is fixed.
GA1DArrayGenome(const unsigned int length,
GAAlleleSet<T> & alleles,
GAObjectiveFunction objective = NULL,
void * userData = NULL,
GA * ga = NULL)
T & allele(const unsigned int i) const
GAAlleleSet<T> & alleles() const
GAAlleleSet<T> & alleles(GAAlleleSet<T> & a)
const T & gene(const unsigned int x=0) const
T & gene(const unsigned int x=0)
T & gene(const unsigned int x, const T & value)
const T & operator[](const unsigned int x) const
T & operator[](const unsigned int x)
int length() const
int length(const int l)
int resize(const int x)
int resizeBehaviour() const
int resizeBehaviour(const unsigned int minx, const int maxx)
void copy(const GA1DArrayGenome<T> & original,
const unsigned int dest, const unsigned int src, const unsigned int length)
void swap(const unsigned int x1, const unsigned int x2)
see also: GAArrayWhen you define an allele set for an array genome, the genome does not make its own copy. Therefore, if you later change the allele set, the array genome will see the changes as well. It will not automatically re-initialize itself.
Use the width and height member functions to get and set the genome dimensions. The gene member function returns the value of the specified element in the array.
The resizeBehaviour function works similarly to that of the 1D array genome. In this case, however, you must also specify for which dimension you are setting the resize behaviour. When resizeBehaviour is called with no arguments, it returns the maximum size if the genome is resizable, or gaNoResize if the size is fixed.
GA2DArrayGenome(const unsigned int width, const unsigned int height,
GAAlleleSet<T> & alleles,
GAObjectiveFunction objective = NULL,
void * userData = NULL,
GA * ga = NULL)
T & allele(const unsigned int i) const
GAAlleleSet<T> & alleles() const
GAAlleleSet<T> & alleles(GAAlleleSet<T> & a)
const T & gene(const unsigned int x, const unsigned int y) const
T & gene(const unsigned int x, const unsigned int y)
T & gene(const unsigned int x, const unsigned int y, const T & value)
int width() const
int width(const int w)
int height() const
int height(const int h)
int resize(const int x, const int y)
int resizeBehaviour(const GADimension which) const
int resizeBehaviour(const GADimension which, const unsigned int min, const int max)
int resizeBehaviour(const unsigned int minx, const int maxx,
const unsigned int miny, const int maxy)
void copy(const GA2DArrayGenome<T> & original,
const unsigned int xdest, const unsigned int ydest,
const unsigned int xsrc, const unsigned int ysrc,
const unsigned int width, const unsigned int height)
void swap(const unsigned int x1, const unsigned int y1,
const unsigned int x2, const unsigned int y2)
see also: GAArrayWhen you define an allele set for an array genome, the genome does not make its own copy. Therefore, if you later change the allele set, the array genome will see the changes as well. It will not automatically re-initialize itself.
Use the width, height, and depth member functions to get and set the genome dimensions. The gene member function returns the value of the specified element in the array.
The resizeBehaviour function works similarly to that of the 1D array genome. In this case, however, you must also specify for which dimension you are setting the resize behaviour. When resizeBehaviour is called with no arguments, it returns the maximum size if the genome is resizable, or gaNoResize if the size is fixed.
GA3DArrayGenome(const unsigned int width, const unsigned int height, const unsigned int depth,
GAAlleleSet<T> & alleles,
GAObjectiveFunction objective = NULL,
void * userData = NULL,
GA * ga = NULL)
T & allele(const unsigned int i) const
GAAlleleSet<T> & alleles() const
GAAlleleSet<T> & alleles(GAAlleleSet<T> & a)
const T & gene(const unsigned int x, const unsigned int y, const unsigned int z) const
T & gene(const unsigned int x, const unsigned int y, const unsigned int z)
T & gene(const unsigned int x, const unsigned int y, const unsigned int z, const T & value)
int width() const
int width(const int w)
int height() const
int height(const int h)
int depth() const
int depth(const int d)
int resize(const int x, const int y, const int z)
int resizeBehaviour(const GADimension which) const
int resizeBehaviour(const GADimension which, const unsigned int min, const int max)
int resizeBehaviour(const unsigned int minx, const int maxx,
const unsigned int miny, const int maxy,
const unsigned int minz, const int maxz)
void copy(const GA3DArrayGenome<T> & original,
const unsigned int xdest, const unsigned int ydest, const unsigned int zdest,
const unsigned int xsrc, const unsigned int ysrc, const unsigned int zsrc,
const unsigned int width, const unsigned int height, const unsigned int depth)
void swap(const unsigned int x1, const unsigned int y1, const unsigned int z1,
const unsigned int x2, const unsigned int y2, const unsigned int z2)
see also: GAArrayCrossover sites are closely coupled with crossover operators. You should use a crossover site that will work with the crossover operator that you specify.
The pick member is called by the genetic algorithm before a crossover occurs. Called with no arguments, the pick method picks at random a site based on the characteristics of the genome to which it belongs. Called with two genomes, pick chooses a crossover site based on the information in the two individuals (assumed to be the parents for a subsequent crossover). Called with a crossover site as its argument, pick copies the specified crossover site.
The mirror member is used when two children are created during mating. Perhaps the simplest example of the use of the mirror function is during crossover on a single dimension binary string. First, the crossover site is selected and stored in the first child. Then the mirror method of the crossover site of the second child is called using the first child's crossover site as its primary 'image'. In the single dimension binary string case, the first child would get the first part of the first parent and the second part of the second parent. The second child would get the first part of the second parent and the second part of the first parent. The site in each parent need not be the same.
GACrossSite() void copy(const GACrossSite &) void pick(GAGenome &) void pick(GAGenome &, const GACrossSite &) void pick(GAGenome&, const GAGenome&, const GAGenome&) void mirror(GAGenome &, const GAGenome &)
The min, max, length, and offset member functions can be used to determine the information about one of the phenotypes. Use the remove member to remove a phenome from the phenotype. The size member returns the number of bits that the set of phenotypes requires.
GABin2DecPhenotype(const unsigned int howmany = 0) void add(const unsigned int nbits, const float min, const float max) void remove(const unsigned int which) int size() const int nPhenotypes() const float min(const unsigned int which) const float max(const unsigned int which) const int length(const unsigned int which) const int offset(const unsigned int which) const
If you call the allele member function with no argument, the allele set picks randomly from the alleles it contains and returns one of them.
For any type, T, the GAAlleleSet<T> has the following member functions:
GAAlleleSet() T & add(const T & allele) T & remove(T & allele) T & allele() const T & allele(const unsigned int i) int size() const
The GAStringAlleleSet is a typedef for the GAlleleSet<char> class.
GAStatistics()
unsigned int populationSize(const unsigned int x)
unsigned int populationSize() const
float pCrossover(const float x)
float pCrossover() const
float pMutation(const float x)
float pMutation() const
unsigned int nGenerations(const unsigned int x)
unsigned int nGenerations() const
float pConvergence(const float x)
float pConvergence() const
unsigned int nConvergence(const unsigned int x)
unsigned int nConvergence() const
unsigned int nBestGenomes()
unsigned int nBestGenomes(const unsigned int n)
int scoreFrequency(const unsigned int x)
int scoreFrequency()
void scoreDestination(const char *filename)
int generation()
float convergence()
int selections()
int mutations()
float online()
float offline()
float bestInitial()
float bestCurrent()
float bestEver()
float bestNth(const unsigned int n = 0)
float score(const unsigned int n)
ostream & scores(ostream &)
GAGenome & bestIndividual(const unsigned int n = 0)
GAPopulation & bestPopulation() const
Here are descriptions of each member function:
- populationSize
- Set/Get the population size. The minimum population size is 1.
- pCrossover
- Set/Get the crossover probability.
- pMutation
- Set/Get the mutation probability.
- nGenerations
- Set/Get the number of generations before terminating the GA.
- pConvergence
- Set/Get the convergence percentage.
- nConvergence
- Set/Get the number of generations to use for the convergence measure. A value of 10 means to use the best-of-generation from 10 generations previous for the convergence test.
- nBestGenomes
- Set/Get the number of unique best genomes to keep.
- scoreFrequency
- Set/Get the frequency at which the best-of-generation should be recorded. A score frequency of 1 means the best-of-generation will be recorded each generation. The default depends on the type of GA that is being used.
- scoreDestination
- Set the name of the file to which the scores should be output. If a filename has been set, scores will be flushed periodically to the specified filename. If you do not want the scores to be automatically written to file, specify NULL as the filename. The default is NULL.
- generation
- Returns the current generation number.
- convergence
- Returns the current convergence measure. This number is the ratio of the Nth previous best-of-generation score to the current best-of-generation score. N is defined by the nConvergence member function.
- selections
- Returns the number of selections made since the GA was initialized.
- mutations
- Returns the number of mutations made since the GA was initialized.
- online
- Returns the average of all scores.
- offline
- Returns the average of the best-of-generation scores.
- bestInitial
- Returns the score of the best individual in the initial population.
- bestCurrent
- Returns the score of the best individual in the current population.
- bestEver
- Returns the score of the best individual ever encountered.
- bestNth
- Returns the score of the best in individual in the Nth previous generation.
- scores
- Print the best-of-generation scores to the specified stream. Output is tab-delimited with each line containing the generation number and the best-of-generation score.
- bestIndividual
- This function returns a reference to the best individual encountered by the genetic algorithm.
- bestPopulation
- This function returns a reference to a population containing the best individuals encountered by the genetic algorithm.
GAParameters(const unsigned int ps = gaDefPopSize,
const float pc = gaDefPCross,
const float pm = gaDefPMut,
const unsigned int ng = gaDefNumGen,
const float pcv = gaDefPConv,
const unsigned int ngv = gaDefNConv,
const unsigned int nbc = gaDefNumBestChrom,
const unsigned int sf = gaDefScoreFrequency1)
unsigned int popSize
float pCross
float pMut
unsigned int nGen
float pConv
unsigned int nConv
unsigned int nBestChrom
unsigned int scoreFreq
The population must always contain at least one individual. If you try to create a population of size 0, it will be created with a size of 1.
You can customize the initialization, evaluation, and sort methods. Use the appropriate member function. Your customized functions must have the appropriate signature.
GAPopulation(const GAGenome & c,
const unsigned int popsize = gaDefPopSize)
GAPopulation(const GAGenome & c,
const GAScalingScheme & scale,
const unsigned int popsize = gaDefPopSize)
int resize(const unsigned int popsize)
int size() const
float sum() const
float ave() const
float var() const
float dev() const
float max() const
float min() const
float fitsum() const
float fitave() const
float fitmax() const
float fitmin() const
GAGenome & best(const unsigned int i = 0) const
GAGenome & worst(const unsigned int i = 0) const
GAGenome & operator[](const unsigned int x) const
GAGenome & individual(const unsigned int x) const
GAGenome * insert(GAGenome *, const unsigned int i)
GAGenome * remove(const unsigned int i)
GAGenome * remove(GAGenome *)
GAGenome * replace(GAGenome *, const int which = gaPopReplaceRandom)
GAGenome * replace(GAGenome *, GAGenome *)
const GAScalingScheme & scaling() const
const GAScalingScheme & scaling(const GAScaling & s)
void evaluate() const
GAPopulationEvaluator evaluator(GAPopulationEvaluator func)
void initialize()
GAPopulationInitializer initializer(GAPopulationInitializer func)
void sort(const GABoolean flag = gaFalse) const
GAPopulationSorter sorter(GAPopulationSorter func)
Here is what each of the member functions does:
- resize
- Change the population to contain the specified number of individuals. If you resize to a larger size, the new individuals will be initialized but not evaluated. If you resize to a smaller size, the best individuals will be kept.
- size
- Returns the number of individuals in the population.
- sum
- Returns the sum of the objective scores.
- ave
- Returns the average of the objective scores.
- var
- Returns the variance of the objective scores.
- dev
- Returns the standard deviation of the objective scores.
- max
- Returns the maximum objective score in the population.
- min
- Returns the minimum objective score in the population.
- fitsum
- Returns the sum of the fitness scores.
- fitave
- Returns the average of the fitness scores.
- fitmax
- Returns the maximum fitness score.
- fitmin
- Returns the minimum fitness score.
- best
- Returns a reference to the best individual in the population.
- worst
- Returns a reference to the worst individual in the population.
- individual, operator[]
- Returns a reference to the specified individual. Indices for individuals in the population start at 0.
- insert
- Insert the specified genome into the population at the specified index.
- remove
- Remove the specified individual from the population. The genome to be replaced can be specified by either an index or by pointer. This function returns a pointer to the genome that was removed from the population. The caller is responsible for the memory used by the returned genome.
- replace
- Replace the specified individual with the first argument. The genome to be replaced can be specified by either an index or by pointer. This function returns a pointer to the genome that was replaced. If no genome was replaced or the specified index or pointer is bogus, it returns NULL.
- scaling
- Set/Get the scaling method for this population.
- evaluate
- Evaluate the population using the method set by the evaluator function. The default evauator simply calls the evaluate member of each genome in the population.
- evaluator
- Specifies which function to use to evaluate the popualation. The specified function must have the proper signature.
- initialize
- Initialize the population using the method set by initializer. The default initializer simply calls the initialize method of each genome in the population.
- initializer
- Specifies which function to use to initialize the popualation. The specified function must have the proper signature.
- sort
- Sort the population using the sorting method set by sorter. If you pass gaTrue to this function, it forces the population to sort.
- sorter
- Specifies which function to use as the sorting method. The specified function must have the proper signature.
void evaluate(const GAPopulation & p) float sum() const float ave() const float max() const float min() const float fitness(const unsigned int i) const float partialSum(const unsigned int i) constThe GA library contains a number of instantiable scaling objects derived from the base class. Here are the creators for these scaling schemes:
GANoScaling() GALinearScaling(const float c = gaDefLinearScalingMultiplier) GASigmaTruncationScaling(const float c = gaDefSigmaTruncationMultiplier) GAPowerLawScaling(const int k = gaDefPowerScalingFactor) GASharing(GADistanceFunction f)
- GA1DBinaryStringGenome
- GA1DBinStrRandomInitializer
- GA1DBinStrSetInitializer
- GA1DBinStrUnsetInitializer
- GA1DBinStrFlipMutator
- GA1DBinStrUniformCrossover
- GA1DBinStr1PtCrossover
- GA1DBinStr2PtCrossover
- GA1DBinStrEvenCrossover
- GA1DBinStrOddCrossover
- GA2DBinaryStringGenome
- GA2DBinStrRandomInitializer
- GA2DBinStrSetInitializer
- GA2DBinStrUnsetInitializer
- GA2DBinStrFlipMutator
- GA2DBinStrUniformCrossover
- GA2DBinStr1PtPadCrossover
- GA2DBinStr1PtClipCrossover
- GA2DBinStrEvenCrossover
- GA2DBinStrOddCrossover
- GA3DBinaryStringGenome
- GA3DBinStrRandomInitializer
- GA3DBinStrSetInitializer
- GA3DBinStrUnsetInitializer
- GA3DBinStrFlipMutator
- GA3DBinStrUniformCrossover
- GA3DBinStr1PtPadCrossover
- GA3DBinStr1PtClipCrossover
- GA3DBinStrEvenCrossover
- GA3DBinStrOddCrossover
The template genomes use the same operator mechanism as the non-template genomes, but because of the template syntax and the design of the GAlib genomes, I cannot assign template functions as default operators for the template objects. You will have to force an instantiation of the operators for the type that you will use.
To do this, you must define the type you will instantiate (using the *_TYPE macro) then include the appropriate header file (ending in .op.h). See the list, tree, or array examples for specific details.
If you want to use variants of these methods, copy them from the source code and modify the <*_TYPE> designation for the type of genome you are going to instantiate. The source code is located be in the files ending in .op.C in the GA include directory.
- GATreeGenome
- GATreeDestructiveMutator
- GATreeSwapSubtreeMutator
- GATreeSwapNodeMutator
- GATree1PtCrossover
- GAListGenome
- GAListDestructiveMutator
- GAListSwapMutator
- GAList1PtCrossover
- GAListPartialMatchCrossover
- GAListOrderCrossover
- GAListCycleCrossover
- GA1DArrayGenome
- GA1DArrayRandomInitializer
- GA1DArrayOrderedInitializer
- GA1DArrayFlipMutator
- GA1DArraySwapMutator
- GA1DArrayUniformCrossover
- GA1DArray1PtCrossover
- GA1DArray2PtCrossover
- GA1DArrayEvenCrossover
- GA1DArrayOddCrossover
- GA1DArrayPartialMatchCrossover
- GA1DArrayOrderCrossover
- GA1DArrayCycleCrossover
- GA2DArrayGenome
- GA2DArrayRandomInitializer
- GA2DArrayFlipMutator
- GA2DArraySwapMutator
- GA2DArrayUniformCrossover
- GA2DArray1PtCrossover
- GA2DArrayEvenCrossover
- GA2DArrayOddCrossover
- GA3DArrayGenome
- GA3DArrayRandomInitializer
- GA3DArrayFlipMutator
- GA3DArraySwapMutator
- GA3DArrayUniformCrossover
- GA3DArray1PtCrossover
- GA3DArrayEvenCrossover
- GA3DArrayOddCrossover
The specializations of template classes come with default methods assigned. You do not have to write your own instance of the methods as you must with the generic template classes. The following methods can be used as you would use the non-template methods listed above.
- GAStringGenome
- GAStringRandomInitializer
- GAStringOrderedInitializer
- GAStringFlipMutator
- GAStringSwapMutator
- GAStringUniformCrossover
- GAString1PtCrossover
- GAString2PtCrossover
- GAStringPartialMatchCrossover
- GAStringOrderCrossover
- GAStringCycleCrossover
 The GAArray object
The GAArray object
The squares are elements in the array. Arrays are 1 dimensional, but derived classes can have 2 or more dimensions. Each element contains a user-specified object.
Any object in the array must have the following methods defined and publicly available:
Here are the creators and member functions for the GAArray<> object:
GAArray(unsigned int s)
GAArray(const GAArray<T> & orig)
GAArray<T> & operator=(const GAArray<T> & orig)
GAArray<T> & operator=(const T array [])
GAArray<T> * clone()
const T & operator[](unsigned int i) const
T & operator[](unsigned int i)
void copy(const GAArray<T> & orig)
void copy(const GAArray<T> & orig, const unsigned int dest,
const unsigned int src, const unsigned int length)
void move(const unsigned int dest,
const unsigned int src, const unsigned int length)
void swap(unsigned int i, unsigned int j)
int size() const
int size(unsigned int n)
int equal(const GAArray<T> & b,
const unsigned int dest, const unsigned int src,
const unsigned int length) const
int notequal(const GAArray<T> & b,
const unsigned int dest, const unsigned int src,
const unsigned int length) const
int operator==(const GAArray<T> & a, const GAArray<T> & b)
int operator!=(const GAArray<T> & a, const GAArray<T> & b)
The elements in an array are indexed starting with 0 (the first element in the array is element number 0). The last element in array with n elements is element n-1.
- clone
- Return a pointer to an exact duplicate of the original array. The caller is responsible for the memory allocated by the call to this function.
- operator[]
- Return a reference to the contents of the ith element of the array.
- copy
- Duplicate the specified array or part of the specified array. If duplicating a part of the specified array, length elements starting at position src in the original are copied into position dest in the copy. If there is not enough space in the copy, the extra elements are not copied.
- move
- Move the number of elements specified with length from position src to position dest.
- swap
- Swap the contents of element i with the contents of element j.
- size
- Return the number of elements in the array.
- equal
- Return 1 if the specified portion of the two arrays is identical, return 0 otherwise.
- notequal
- Return 1 if the specified portion of the two arrays is not identical, return 0 otherwise.
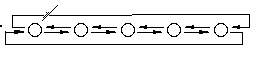 The GAList object
The GAList object
The circles are nodes in the list. Each node contains a user-specified object; the initialization method determines the size of the list and the contents of each node. The list is circular and doubly linked.
The template-ized GAList<T> is derived from a generic list base class called GAListBASE. The template list is defined in listtmpl.h, the list base class is defined in listbase.h
Any object used in the nodes must have the following methods defined and publicly available:
Here are the creators and member functions for the GAList<> object:
GAList()
GAList(const T & t)
GAList(const GAList<T> & orig)
GAList<T> & operator=(const GAList<T> & orig)
GAList<T> * clone()
void copy(const GAList<T> & orig)
void destroy()
void swap(const unsigned int, const unsigned int)
T * remove()
void insert(GAList<T> * t, GAListBASE::Location where=AFTER)
void insert(const T & t, GAListBASE::Location where=AFTER)
T * current()
T * head()
T * tail()
T * next()
T * prev()
T * warp(const unsigned int i)
T * warp(const GAListIter<T> & i)
T * operator[](const unsigned int i)
int size() const
Each list object contains an iterator. The list's traversal member functions (next, prev, etc) simply call the member functions on the internal iterator. You can also instantiate iterators external to the list object so that you can traverse the list without modifying its state. The list iterator has the following member functions:
GAListIter(const GAList<T> &) T * current() T * head() T * tail() T * next() T * prev() T * warp(const GAList<T> & t) T * warp(const GAListIter<T> & i) T * warp(const unsigned int i)The list base class defines the following constants for specifying where insertions should take place (these are relative to the node to which the iterator is currently pointing):
GAListBASE::HEAD GAListBASE::TAIL GAListBASE::BEFORE GAListBASE::AFTERNodes in the list are numbered from 0 to 1 less than the list size. The head node is node 0.
These functions change the state of the list.
These functions do not change the contents of the list, but they change the state of the list's internal iterator.
- clone
- Return a pointer to an exact duplicate of the original list. The caller is responsible for the memory allocated by the call to this function.
- copy
- Duplicate the specified list.
- destroy
- Destroy the current node in the list. This function uses the location of the internal iterator to determine which node should be destroyed. If the head node is destroyed, the next node in the list becomes the head node.
- remove
- Returns a pointer to the contents of the current node and removes the current node from the list. The iterator moves to the previous node. The caller is responsible for the memory used by the contents.
- swap
- Swap the positions of the two specified nodes.
- insert
- Add a node or list to the list. The insertion is made relative to the location of the internal iterator. The where flag specifies whether the insertion should be made before or after the current node.
- current
- Returns a pointer to the contents of the current node.
- head
- Returns a pointer to the contents of the first node in the list.
- tail
- Returns a pointer to the contents of the last node in the list.
- next
- Returns a pointer to the contents of the next node.
- prev
- Returns a pointer to the contents of the previous node.
- warp
- Returns a pointer to the contents of the ith node in the list, or a pointer to the element in the list pointed to by the specified iterator.
- operator[]
- Returns a pointer to the contents of the ith node in the list (same as warp).
When you do an insertion, the list makes a copy of the specified object (allocating space for it in the process). The internal iterator is left pointing to the node which was just inserted. The insertion function uses the copy constructor member to do this, so the objects in your list must have a copy constructor defined. The new node is inserted relative to the current location of the list's internal iterator. Use the where flag to determine whether the new node will be inserted before or after the current node, or if the new node should become the head node of the list.
The remove member returns a pointer to the object that was in the specified node. You are responsible for deallocating the memory for this object! The destroy member deallocates the memory used by the object in the current node. In both cases the iterator is left pointing to the node previous to the one that was deleted.
The swap member swaps the indicated nodes. The internal interator is not affected. If the iterator was pointing to one of the nodes before the swap it will still point to that node after the swap, even if that node was swapped.
The warp method moves the iterator to the specified node in the list. The head node is number 0.
All of the list traversal functions (prev, next, current, etc) return a pointer to the contents of the node on which they are operating. You should test the pointer to see if it is NULL before you dereference it. When you call any of the traversal functions, the list's internal iterator is left pointing to the node to which traversal function moved. You can create additional iterators (external to the list) to keep track of multiple positions in the list.
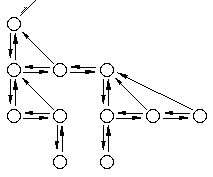 The GATree object
The GATree object
The circles are nodes in the tree. Each node contains a user-specified object; the initialization method determines the tree topology and the contents of each node. Each tree contains one (and only one) root node. Each level in the tree is a circular, doubly linked list. The head of each list is called the 'eldest' child, each node in a level has a link to its parent, and each parent has a link to the eldest of its children (if it has any children).
The template-ized GATree<T> is derived from a generic tree base class called GATreeBASE. The template tree is defined in treetmpl.h, the tree base class is defined in treebase.h
Any object used in the nodes have the following methods defined and publicly available:
Here are the creators and member functions for the GATree<> object:
GATree()
GATree(const T & t)
GATree(const GATree<T> & orig)
GATree<T> & operator=(const GATree<T> & orig)
GATree<T> * clone()
void copy(const GATree<T> & orig)
void destroy()
void swaptree(GATree<T> * t)
void swaptree(const unsigned int, const unsigned int)
void swap(const unsigned int, const unsigned int)
GATree<T> * remove()
void insert(GATree<T> * t, GATreeBASE::Location where=BELOW)
void insert(const T & t, GATreeBASE::Location where=BELOW)
T * current()
T * root()
T * next()
T * prev()
T * parent()
T * child()
T * eldest()
T * youngest()
T * warp(const unsigned int i)
T * warp(const GATreeIter<T> & i)
int ancestral(const unsigned int i, const unsigned int j) const
int size()
int depth()
int nchildren()
int nsiblings()
Each tree object contains an iterator. The tree's traversal member functions (next, prev, etc) simply call the member functions on the internal iterator. You can also instantiate iterators external to the tree object so that you can traverse the tree without modifying its contents. The tree iterator has the following member functions:
GATreeIter(const GATree<T> & t)
T * current()
T * root()
T * next()
T * prev()
T * parent()
T * child()
T * eldest()
T * youngest()
T * warp(const GATree<T> & t)
T * warp(const GATreeIter<T> & i)
T * warp(const unsigned int i)
int size()
int depth()
int nchildren()
int nsiblings()
The tree base class defines the following constants for specifying where insertions should occur (these are relative to the node to which the iterator is currently pointing):
GATreeBASE::ROOT GATreeBASE::BEFORE GATreeBASE::AFTER GATreeBASE::BELOWNodes in a tree are numbered starting at 0 then increasing in a depth-first traversal of the tree. The root node is node 0. A tree can have only one root node, but any node in the tree can have any number of children.
These functions change the state of the tree.
These functions do not change the contents of the tree, but they change the state of the tree's internal iterator.
- clone
- Return a pointer to an exact duplicate of the original tree. The caller is responsible for the memory allocated by the call to this function.
- copy
- Duplicate the specified tree.
- destroy
- Destroy the current node in the tree. If the node has children, the entire sub-tree connected to the node is destroyed as well. This function uses the location of the internal iterator to determine which node should be destroyed. If the root node is destroyed, the entire contents of the tree will be destroyed, but the tree object itself will not be deleted.
- remove
- Returns a pointer to a new tree object whose root node is the (formerly) current node of the original tree. Any subtree connected to the node stays with the node. The iterator moves to the previous node in the current generation, or the parent node if no elder sibling exists. The caller is responsible for the memory used by the new tree.
- swap
- Swap the contents of the two specified nodes. Sub-trees connected to either node are not affected; only the specified nodes are swapped.
- swaptree
- Swap the contents of the two specified nodes as well as any sub-trees connected to the specified nodes.
- insert
- Add a node or tree to the tree. The insertion is made relative to the location of the internal iterator. The where flag specifies whether the insertion should be made before, after, or below the current node.
- current
- Returns a pointer to the contents of the current node.
- root
- Returns a pointer to the contents of the root node of the tree.
- next
- Returns a pointer to the contents of the next node in the current generation.
- prev
- Returns a pointer to the contents of the previous node in the current generation.
- parent
- Returns a pointer to the contents of the parent of the current node. If the current node is the root node, this function returns NULL.
- child
- Returns a pointer to the contents of the eldest child of the current node. If the current node has no children, this function returns NULL.
- eldest
- Returns a pointer to the contents of the eldest node in the current generation. The eldest node is the node pointed to by the 'child' function in the node's parent.
- youngest
- Returns a pointer to the contents of the youngest node in the current generation.
- warp
- Returns a pointer to the contents of the ith node in the tree, or a pointer to the element in the tree pointed to by the specified iterator.
- size
- Returns the number of nodes in the tree. When called as the member function of a tree iterator, this function returns the size of the subtree connected to the iterator's current node.
- depth
- Returns the number of generations (the depth) of the tree. When called as the member function of a tree iterator, this function returns the depth of the subtree connected to the iterator's current node.
- nchildren
- Returns the number of children of the current node.
- nsiblings
- Returns the number of nodes in the current generation.
- ancestral
- Returns 1 if one of the two specified nodes is the ancestor of the other, returns 0 otherwise.
When you do an insertion, the tree makes a copy of the specified object (allocating space for it in the process). The internal iterator is left pointing to the node which was just inserted. The insertion function uses the copy constructor member to do this, so the objects in your tree must have a copy constructor defined. The new node is inserted relative to the current location of the tree's internal iterator. Use the where flag to determine whether the new node will be inserted before, after, or below the current node, or if the new node should become the root node of the tree.
The remove member returns a pointer to a tree. The root node of this tree is the node at which the iterator was pointing. You are responsible for deallocating the memory for this tree! The destroy member deallocates the memory used by the object in the current node and completely destroys any subtree hanging on that node. In both cases, the iterator is left pointing to the elder child or parent of the node that was removed/destroyed.
The nsiblings method returns the number of children in the level of the node to which the iterator is pointing. The nchildren method returns the number of children of the node to which the iterator in pointing.
The warp method moves the iterator to the specified node in the tree. The head node is number 0 then the count increases as a depth-first traversal of the tree.
All of the tree traversal functions (prev, next, current, etc) return a pointer to the contents of the node on which they are operating. You should test the pointer to see if it is NULL before you dereference it. Also, the iterator is left pointing to the node to which you traverse with each traversal function. You can create additional iterators (external to the tree) to keep track of multiple positions in the tree.
GABoolean GATerminateUponGeneration(GA & ga){
return(ga.generation() < ga.nGenerations() ? gaFalse : gaTrue);
}
The second example compares the score of the best individual in the current population with the average score in the current population. If the ratio of these exceeds a specified threshhold, it returns gaTrue to signify that the GA should stop. Basically this means that the entire population has converged to a 'good' score.
const float desiredRatio = 0.95; // stop when pop average is 95% of best
GABoolean GATerminateUponScoreConvergence(GA & ga){
if(ga.population().ave()/ga.population().best().score() > desiredRatio)
return gaTrue;
else
return gaFalse;
}
GAGenome &
GARouletteWheelSelector(const GAPopulation & p)
{
static float cutoff;
static int i, upper, lower;
cutoff = GARandomFloat() * p.scaling().sum();
lower = 0; upper = p.size()-1;
while(upper >= lower){
i = lower + (upper-lower)/2;
if(p.scaling().partialSum(i) > cutoff)
upper = i-1;
else
lower = i+1;
}
lower = Min(p.size()-1, lower);
lower = Max(0, lower);
return(p.scaling().individual(lower));
}
class GASigmaTruncationScaling : public GAScalingScheme {
GADefineIdentity("GASigmaTruncationScaling", 18):
public:
GASigmaTruncationScaling(const float m=gaDefSigmaTruncationMultiplier) :
GAScalingScheme(gaDefSize){multiplier(m);}
GASigmaTruncationScaling(const GAPopulation & p,
const float m=gaDefSigmaTruncationMultiplier) :
GAScalingScheme(p){multiplier(m);}
GASigmaTruncationScaling(const GASigmaTruncationScaling & arg) :
GAScalingScheme(arg.n){copy(arg);}
GASigmaTruncationScaling & operator=(const GAScalingScheme & arg)
{copy(arg); return(*this);}
GASigmaTruncationScaling & operator=(const GASigmaTruncationScaling & arg)
{copy(arg); return(*this);}
virtual ~GASigmaTruncationScaling(){}
void copy(const GAScalingScheme & arg){
if(&arg != this && sameClass(arg))
GAScalingScheme::copy(arg); c=((GASigmaTruncationScaling&)arg).c;
}
GAScalingScheme * clone() const {return new GASigmaTruncationScaling(*this);}
void evaluate(const GAPopulation & p);
float multiplier(const float fm);
float multiplier() const {return c;}
protected:
float c; // std deviation multiplier
};
void
GASigmaTruncationScaling::evaluate(const GAPopulation & p)
{
if(evaluated == gaTrue) return;
memcpy(ind, p.ind, n*sizeof(GAGenome*));
for(int i=0; i<n; i++){
fit[i] = ind[i]->score() - p.ave() + c * p.dev();
if(fit[i] < 0) fit[i] = 0;
}
GAQuickSort(ind, fit, 0, n-1);
fitMin = fitMax = fitSum = psum[0] = fit[0];
for(i=1; i<n; i++){
fitSum += fit[i];
psum[i] = fitSum;
if(fit[i] > fitMax) fitMax = fit[i];
if(fit[i] < fitMin) fitMin = fit[i];
}
fitAve = fitSum/n;
evaluated = gaTrue;
}
Here is the implementation of an initializer for the GATreeGenome<int> class.
void
TreeInitializer(GAGenome & c)
{
GATreeGenome<int> &child=(GATreeGenome<int> &)c;
// destroy any pre-existing tree
child.root();
child.destroy();
// Create a new tree with depth of 'depth' and each eldest node containing
// 'n' children (the other siblings have none).
int depth=2, n=3, count=0;
child.insert(count++,GATreeBASE::ROOT);
for(int i=0; i<depth; i++){
child.eldest();
child.insert(count++);
for(int j=0; j<n; j++)
child.insert(count++,GATreeBASE::AFTER);
}
}
The crossover site should have been selected before the crossover method is called. The crossover method should check the site and pick a site if one has not been selected already. Each genome contains a crossover site. The child's site should be used by the crossover operator.
Here is the implementation of the partial match crossover operator for the GAListGenome<char> class.
void
GAListPartialMatchCrossover(GAGenome &c, const GAGenome &a, const GAGenome &b)
{
GAListGenome<char> &child=(GAListGenome<char> &)c;
GAListGenome<char> &mom=(GAListGenome<char> &)a;
GAListGenome<char> &dad=(GAListGenome<char> &)b;
GAListOrderedCrossSite<char> & xsite =
(GAListOrderedCrossSite<char> &)child.crossoverSite();
if(xsite.a == gaNoSite || xsite.b == gaNoSite) xsite.pick(c);
child.GAList<char>::copy(mom);
GAListIter<char> iter(dad);
char *index = iter.warp(xsite.a);
for(int i=xsite.a; i<xsite.b; i++, index=iter.next()){
if(*child.head() == *index){
child.swap(i,0);
}
else{
int j;
for(j=1; (j<child.size()) && (*child.next() != *index); j++);
child.swap(i,j); // no op if j>size
}
}
child.head(); // set iterator to head of list
}
Here is the implementation of the flip mutator for the GA1DBinaryString class.
int
GA1DBinStrFlipMutator(GAGenome & c, float pmut)
{
GA1DBinaryStringGenome &child=(GA1DBinaryStringGenome &)c;
register int n, i;
if(pmut <= 0.0) return(0);
float nMut = pmut * (float)(child.length());
if(nMut < 1.0){ // we have to do a flip test on each bit
nMut = 0;
for(i=child.length()-1; i>=0; i--){
if(GAFlipCoin(pmut)){
child.gene(i, ((child.gene(i) == 0) ? 1 : 0));
nMut++;
}
}
}
else{ // only flip the number of bits we need to flip
for(n=0; n<nMut; n++){
i = GARandomInt(0, child.length()-1); // the index of the bit to flip
child.gene(i, ((child.gene(i) == 0) ? 1 : 0));
}
}
return((int)nMut);
}